Copper homeostasis networks in the bacterium Pseudomonas aeruginosa.
Abstract
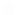
Bacterial copper (Cu<sup>+</sup>) homeostasis enables both precise metallation of diverse cuproproteins and control of variable metal levels. To this end, protein networks mobilize Cu<sup>+</sup> to cellular targets with remarkable specificity. However, the understanding of these processes is rather fragmented. Here, we use genome-wide transcriptomic analysis by RNA-Seq to characterize the response of <i>Pseudomonas aeruginosa</i> to external 0.5 mm CuSO<sub>4</sub>, a condition that did not generate pleiotropic effects. Pre-steady-state (5-min) and steady-state (2-h) Cu<sup>+</sup> fluxes resulted in distinct transcriptome landscapes. Cells quickly responded to Cu<sup>2+</sup> stress by slowing down metabolism. This was restored once steady state was reached. Specific Cu<sup>+</sup> homeostasis genes were strongly regulated in both conditions. Our system-wide analysis revealed induction of three Cu<sup>+</sup> efflux systems (a P<sub>1B</sub>-ATPase, a porin, and a resistance-nodulation-division (RND) system) and of a putative Cu<sup>+</sup>-binding periplasmic chaperone and the unusual presence of two cytoplasmic CopZ proteins. Both CopZ chaperones could bind Cu<sup>+</sup> with high affinity. Importantly, novel transmembrane transporters probably mediating Cu<sup>+</sup> influx were among those largely repressed upon Cu<sup>+</sup> stress. Compartmental Cu<sup>+</sup> levels appear independently controlled; the cytoplasmic Cu<sup>+</sup> sensor CueR controls cytoplasmic chaperones and plasma membrane transporters, whereas CopR/S responds to periplasmic Cu<sup>+</sup> Analysis of Δ<i>copR</i> and Δ<i>cueR</i> mutant strains revealed a CopR regulon composed of genes involved in periplasmic Cu<sup>+</sup> homeostasis and its putative DNA recognition sequence. In conclusion, our study establishes a system-wide model of a network of sensors/regulators, soluble chaperones, and influx/efflux transporters that control the Cu<sup>+</sup> levels in <i>P. aeruginosa</i> compartments.
Description
This work was supported by NIGMS, National Institutes of Health Grant R01 GM114949 (to J. M. A.).
Collections
- Artículos de Revista [4552]